The Unconventional Key: CIRBP’s Phosphorylation-Free Nuclear Entry
Unlocking the Nuclear Door: A Different Kind of Key
You know, cells are like tiny, bustling cities, and the nucleus is the city hall – the control center where all the important genetic blueprints are kept. Getting proteins into and out of this city hall is a massive operation, handled by specialized transport systems. One of the key players in this cellular logistics game is a protein called Transportin 3, or TNPO3. It’s like a dedicated shuttle service for getting certain proteins across the nuclear border, through something called the nuclear pore complex.
For a long time, we thought we had a pretty good handle on how TNPO3 worked, especially with its favorite passengers, the SRSF proteins. These guys often have a special “boarding pass” called a nuclear localization signal (NLS), typically rich in arginine-serine (SR/RS) repeats. The general wisdom was that for TNPO3 to really grab onto these passengers and shuttle them in, those serines often needed a little chemical tag – a phosphate group – added by specific kinases. It was like needing your ticket stamped before you could board.
But science, bless its curious heart, always throws us curveballs! Recent studies started spotting proteins hitching a ride with TNPO3 that didn’t seem to have these classic SR/RS repeats. One protein that popped up was the cold-inducible RNA-binding protein, or CIRBP. Now, CIRBP is a fascinating character itself, involved in things like stabilizing mRNA and helping with DNA repair. It hangs out in the nucleus but can zip out to the cytoplasm under stress, forming these cool structures called stress granules. Its location is super important for what it does, influencing cell growth and even playing a role in cancer.
CIRBP has a region that looked like it *could* be an NLS, but it was a bit different – an RSY-NLS, rich in arginine, serine, *and* tyrosine. And here’s the kicker: initial hints suggested it didn’t need that phosphorylation stamp to get into the nucleus via TNPO3. This was a real head-scratcher! How was TNPO3 recognizing this different signal? That’s where our journey in this particular study really began – we wanted to crack the code of this unconventional interaction.
Peeking Inside: The Structural Secrets
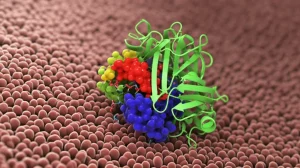
To figure out how TNPO3 was binding to CIRBP without the usual phosphorylation signal, we needed to get up close and personal. The best way to do that is to look at their structures when they’re bound together. So, we turned to a powerful technique called X-ray crystallography. Imagine shining incredibly bright X-rays through a tiny, perfect crystal made of our TNPO3-CIRBP complex. The way the X-rays bounce off the atoms gives us clues to build a 3D map of the molecules.
And oh boy, did that map (which you can find in the Protein Data Bank as PDB: 8CMK) reveal some cool stuff! We saw that the RSY region of CIRBP snuggles right into a spot on TNPO3, specifically between its HEAT repeats 14 and 17. But the way it binds is totally different from the classic SR/RS passengers. Instead of just a floppy chain, the CIRBP peptide forms two small helical turns when it binds. It’s like it changes shape just for the occasion, and TNPO3 even opens up a bit to accommodate it.
The real stars of the show in this interaction turned out to be the tyrosine residues within the CIRBP RSY-NLS. Specifically, three tyrosines (Y160, Y164, and Y167) were absolutely crucial. They weren’t just sitting there; they were actively involved in a whole network of interactions with TNPO3. We saw things like hydrogen bonds, where atoms share a tiny attraction, and even something called pi stacking and cation-pi interactions, which are special ways that aromatic rings (like those in tyrosine) and positively charged groups can interact. It’s like a molecular handshake, but with multiple hands!
These tyrosine interactions seemed to be doing the heavy lifting, providing the necessary grip for TNPO3 to bind CIRBP tightly. This is a big deal because, in the classic SR/RS NLSs, it’s the negatively charged phosphate group on the serine that often makes those crucial contacts with positively charged arginines on TNPO3. Here, the tyrosines, with their slightly polar -OH groups and their aromatic rings, were creating a different kind of interaction landscape. TNPO3 even has a little cavity near a key tyrosine (Y702) that seems perfectly shaped to grab onto CIRBP’s tyrosines. It’s a clever workaround by nature to achieve binding without relying on phosphorylation.

The Unexpected Twist: Phosphorylation Inhibits Binding!
Now, remember how we said phosphorylation is usually *required* or *enhances* TNPO3 binding for classic NLSs? Well, here’s where the CIRBP story gets really surprising. We decided to test what happens if CIRBP *does* get phosphorylated in its RSY region. Using techniques like NMR spectroscopy and ITC (Isothermal Titration Calorimetry), we measured how strongly phosphorylated CIRBP binds to TNPO3 compared to the unphosphorylated version.
The results were almost the opposite of what you’d expect based on the classic model! Phosphorylation of certain serines (S146, S148, S152) in the broader CIRBP region significantly *decreased* TNPO3 binding affinity – by about tenfold! And when we looked at phosphorylating the key tyrosines (Y164, Y167) or a nearby serine (S166) within the core binding motif itself using synthetic peptides, the binding was practically abolished. It seems that adding a phosphate group to these specific spots on CIRBP actually *gets in the way* of TNPO3 binding.
We confirmed this surprising finding in living cells using a clever trick where we could control when CIRBP is released into the cytoplasm and watch if it goes into the nucleus. Mutating just one of those key tyrosines (Y164A) or the crucial arginine (R161A) completely messed up CIRBP’s nuclear import. This matches our binding data perfectly – if it can’t bind TNPO3 properly, it can’t get into the nucleus efficiently.
This tells us something really important: for CIRBP, phosphorylation acts as a *negative* regulator of nuclear import by TNPO3. This is a stark contrast to the positive regulation seen with classical SR/RS NLSs. It suggests there might be a whole different class of TNPO3 cargoes out there, regulated in a completely opposite way. We don’t yet know exactly which kinase enzymes are responsible for phosphoryl phosphorylating these specific serines and tyrosines on CIRBP, but identifying them will be a key piece of the puzzle in understanding how CIRBP’s nuclear entry is controlled.
Searching for More Unconventional Passengers
Finding this unique phosphorylation-independent, tyrosine-driven mechanism in CIRBP got us thinking: are there other proteins out there using a similar trick to get into the nucleus via TNPO3? We used computational tools to scan the human proteome for sequences that look like the CIRBP RSY-NLS motif (Y-R-x-S-Y-D-S-Y or simplified versions like Y-R-x(2,3)-Y-x(2,3)-Y, even including phenylalanine and tryptophan as alternatives to tyrosine).
Our search turned up a bunch of potential candidates – over 90 proteins with the simplified tyrosine-rich motif, many located in intrinsically disordered regions (floppy parts of proteins) where NLSs are often found. We even found some proteins that weren’t previously known to be TNPO3 cargoes but have this motif, like RBMX. We tested a few of these candidates using NMR and computational predictions (like AlphaFold2) and found that several of them indeed seem to interact directly with TNPO3, often binding to the same spot as CIRBP.

This suggests that the CIRBP RSY-NLS isn’t just a one-off anomaly. There’s likely a whole family of proteins that use these tyrosine-rich motifs to get into the nucleus via TNPO3, bypassing the need for phosphorylation. Some of these proteins are already known to be nuclear, which makes sense, while others might be new, undiscovered TNPO3 passengers. Finding these new potential cargoes opens up exciting avenues for future research to understand their biological roles and how their nuclear transport is regulated.
Why This Matters
So, why is understanding this unconventional nuclear import mechanism so important? Well, nuclear transport isn’t just about moving proteins around; it’s fundamental to how cells function. Proteins that regulate gene expression, RNA processing, and DNA repair need to be in the nucleus to do their jobs. If their transport is disrupted, it can lead to all sorts of problems, including developmental defects and diseases like muscular dystrophy, which is linked to TNPO3 mutations.
Discovering that TNPO3 uses a completely different strategy to import proteins like CIRBP – one that relies on tyrosines instead of phosphorylated serines and is *inhibited* by phosphorylation rather than promoted – changes our understanding of this crucial transporter. It adds another layer of complexity and regulation to the nuclear import landscape. This alternative mechanism could be particularly important for proteins whose function or localization needs to be rapidly switched based on cellular signals, perhaps involving those inhibitory phosphorylation events.
Think about CIRBP itself. Its movement between the nucleus and cytoplasm is tied to stress responses. This newly discovered negative regulation by phosphorylation could be a way for the cell to quickly keep CIRBP out of the nucleus under specific conditions, perhaps when those inhibitory kinases are active. It’s a sophisticated switch controlling where this important protein can operate.
Wrapping It Up
In a nutshell, this study peeled back the layers on how TNPO3 interacts with CIRBP and found something truly unexpected. It’s not the classic phosphorylation-dependent handshake; it’s a unique grip mediated by tyrosines. And even more surprisingly, phosphorylation of CIRBP’s NLS acts like a molecular “stop” sign, preventing TNPO3 binding and nuclear import. This reveals a fascinating alternative mechanism for nuclear transport by TNPO3 and hints at a whole new class of cargo proteins that might be using similar tyrosine-rich signals.
There’s still plenty to explore, of course. We need to confirm the binding and biological significance of all those potential new cargoes we identified. And finding the specific kinases that put those inhibitory phosphate tags on CIRBP’s tyrosines and serines is a big next step. But for now, it’s pretty cool to know that even in well-studied processes like nuclear import, there are still unconventional secrets waiting to be discovered, showing just how clever and adaptable our cells really are.

Source: Springer