AI Unlocks Back Pain Secrets: Segmenting Fascia with Ultrasound
Okay, let’s talk about something that messes with a lot of us: low back pain. You know, that nagging ache that just won’t quit? For ages, we thought about bones, muscles, nerves… but only recently have we really started to appreciate the role of this cool, often-overlooked structure called the thoracolumbar fascia (TLF).
Turns out, this layered connective tissue in your back isn’t just passive packing material; it’s actively involved in low back pain (LBP). This is a big deal because understanding it better could totally change how we treat LBP.
The Problem with Getting a Good Look
So, if the fascia is important, how do we check it out? Well, ultrasound is a fantastic tool because it’s non-invasive and lets us see things in real-time. But here’s the catch: getting a reliable ultrasound image of the TLF and knowing exactly what you’re looking at is surprisingly tricky. It depends heavily on the specific ultrasound machine settings and, honestly, how experienced the person holding the probe is. It’s like trying to find a specific thread in a complex tapestry – the clarity varies wildly.
This lack of a clear, objective way to evaluate the fascia during a standard clinical visit has created a bit of a gap. It makes it hard for clinicians to really see what’s going on with the TLF and how it might be contributing to someone’s pain.
Enter Deep Learning: Our Digital Assistant
That’s where we thought, “Hey, what if we could get a computer to help?” Our goal with this work was to see if we could use a fancy type of artificial intelligence called deep learning to automatically identify and segment (basically, draw a precise outline around) the thoracolumbar fascia in ultrasound images. We wanted to fill that gap and make TLF evaluation more objective and accessible.
Building the Brain: Our Approach
To do this, we needed data – lots of it! We gathered a total of 538 ultrasound images of the TLF from people who were experiencing LBP. We then used these images to train and test a deep learning network. Think of training like showing the computer hundreds of examples, saying, “See this outline? That’s the fascia!”
We used a well-known architecture for this kind of task called a U-Net. It’s proven to be pretty effective for segmenting medical images, especially musculoskeletal structures in ultrasound.
We split our main dataset (let’s call it Test set 1) into parts for training, validation (checking the model’s learning as it goes), and final testing (images the model had never seen). But we didn’t stop there. To really push the boundaries and see if our approach was generalizable – meaning it would work even if the ultrasound machine, the operator, or even the patient group was different – we collected an *additional* test set (Test set 2) from a completely different clinical center.
Before feeding the images into the network, we did some standard cleanup: cropping the interesting part, resizing everything to a consistent size (256×256 pixels), and normalizing the image data. We also had to meticulously label the images, outlining the TLF itself, the tissue above it, and the tissue below it (including the epimysial fascia covering the muscles).
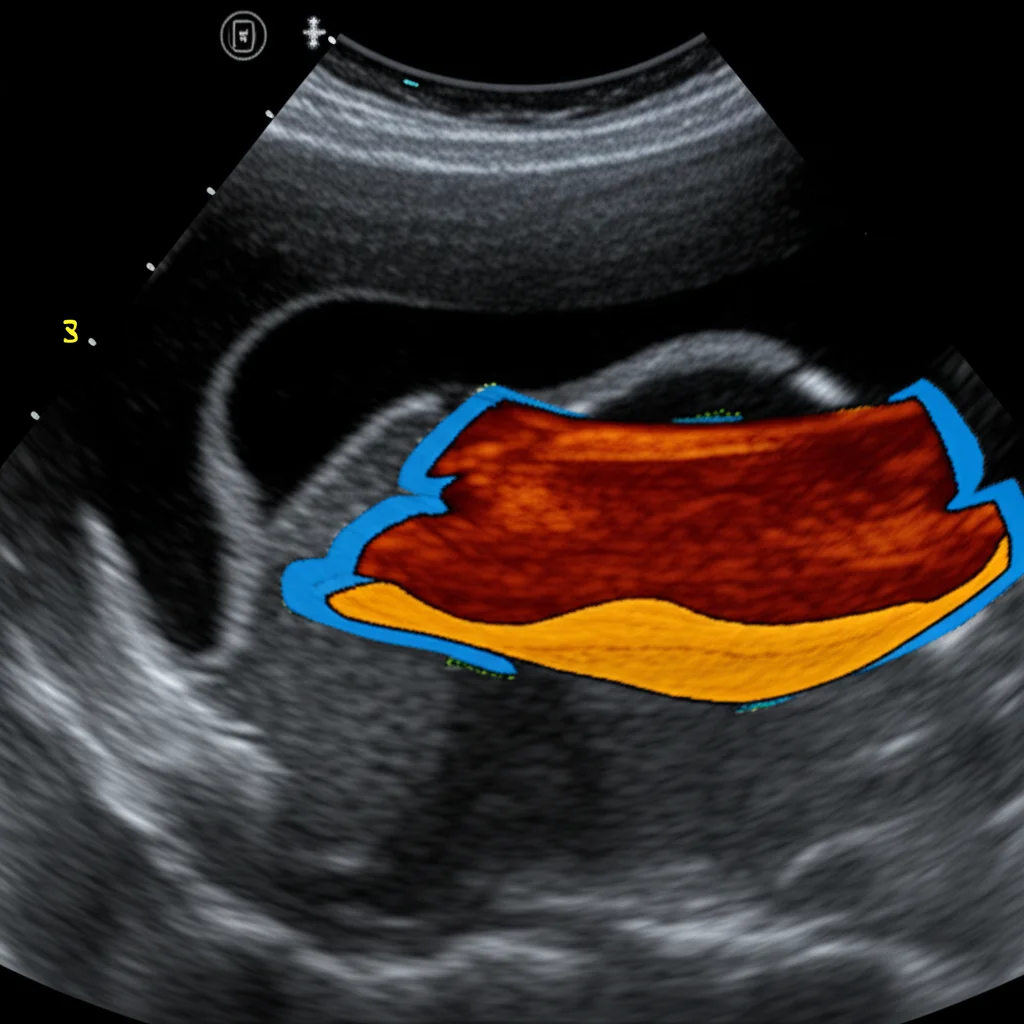
The Results: Pretty Impressive!
So, how did our digital assistant perform? The U-Net model learned remarkably well, achieving a final training accuracy of 0.99 and a validation accuracy of 0.91. When we tested it on the unseen images from Test set 1, the prediction accuracy was 0.94. We also looked at other standard metrics like the mean Intersection over Union (IoU) index (0.82) and the Dice-score (0.76), which measure how well the predicted outline matches the actual outline.
Here’s where it gets really interesting: on Test set 2, the one collected from a totally different setup, the results were even *better*! We saw a mean IoU of 0.85 and a Dice-score of 0.91. This was fantastic news because it strongly suggests the method isn’t just specific to one lab or one machine.
And because numbers only tell part of the story, we had two expert clinicians visually inspect the segmentations produced by our model. They confirmed that the predictions were valid and accurate.
Another bonus? The model was fast. Once trained, segmenting a new image could be done almost in real-time, taking just seconds.
Why This Matters for You (and Your Back)
This isn’t just a cool tech demo; it has real implications for clinical practice. Right now, investigating the TLF often involves expensive imaging like MRIs, which still don’t always give the detailed, real-time view that ultrasound can. But as we discussed, manual ultrasound evaluation is subjective and operator-dependent.
Our automated segmentation tool changes the game. It offers an objective, rapid, and real-time way to identify and potentially quantify changes in the thoracolumbar fascia. Imagine a clinician being able to quickly get a precise outline of the fascia during a standard ultrasound scan! This could help them:
- Identify if the fascia is thicker than expected (which is associated with LBP).
- Assess the gliding movement between the fascial layers (reduced gliding is also linked to chronic LBP).
- Track changes in the fascia over time or after treatment.
- Target specific areas of the fascia during physical therapy or other interventions.
This could lead to more personalized and effective treatments for low back pain, potentially reducing costs and improving patient outcomes. It helps turn ultrasound into a more powerful diagnostic and assessment tool for this crucial structure.
Looking Ahead: What’s Next?
While we’re super excited about these results, this is just the beginning. We used a standard U-Net because we wanted to prove the concept was feasible. Future work could involve comparing it to other cutting-edge AI models to see if we can push the performance even further.
Data is always key. We need even more varied data, perhaps including images with different types of noise or from patients with specific, complex conditions, to make the tool even more robust. Also, developing better ways to do data augmentation for fascial structures is important – you want to make the model see variations without creating unrealistic anatomy.
Moving from static images to segmenting fascia in real-time ultrasound *videos* is the next big step, as that’s how ultrasound is often used clinically. We also want to develop automated tools to quantify things like fascial thickness and gliding directly from the segmented images.
The Takeaway
In a nutshell, our study shows that using deep learning to automatically segment the thoracolumbar fascia from ultrasound images is not only possible but shows really promising results, especially with its ability to work across different clinical setups. This tool has the potential to significantly improve how we investigate the fascia’s role in low back pain and other musculoskeletal conditions, paving the way for more objective assessments and personalized treatments. It’s a pretty exciting step forward in bringing AI into the clinic to help us understand and treat pain better!
Source: Springer