Cracking the Code: Making AI Explainable for AMD Diagnosis
Hey there! Let’s chat about something super important in the world of medicine and tech: making AI understandable. You see, deep learning models are becoming rockstars at spotting diseases from medical images, like those cool pictures of the back of your eye (fundus images). They’re getting really good, sometimes even better than traditional methods! But here’s the catch, and it’s a big one: they’re often like a total “black box.” You feed them an image, they give you a diagnosis, but they don’t tell you *why* they think that. For doctors and patients, that lack of transparency is a major hurdle. It’s tough to trust a diagnosis if you don’t understand the reasoning behind it, right?
The AI Black Box Problem in Healthcare
This whole “black box” thing is a real barrier to getting AI widely used in clinics. We need to move beyond just saying, “The AI says you have X.” We need the AI to say, “I think you have X because I see Y and Z features in this image.” That’s where the field of explainable AI (XAI) comes in. A common trick in XAI for images is using heatmaps, like Grad-CAM, to show which parts of the image the model focused on. It’s a start, but honestly, it doesn’t give us the full picture, especially in medicine.
Here’s the core issue: Do those highlighted regions on the heatmap actually correspond to things a doctor would look for? Can the model back up its diagnosis with real medical reasoning? Most existing XAI methods give you algorithmic explainability – they show you how the algorithm works internally. But what we really need is medical explainability – the ability for the model to justify its diagnosis based on medical knowledge. That’s a whole different ballgame, and it’s surprisingly underexplored.
Building trust is key. Doctors and patients need to feel confident in AI-based diagnoses. And that confidence comes from understanding. Our work dives headfirst into this challenge, specifically focusing on age-related macular degeneration (AMD).
Why AMD? And Why Explainability Matters Here
AMD is a big deal. It’s a leading cause of vision loss for folks over 50, affecting millions worldwide. Early detection is crucial because the vision loss is irreversible, and treatments work best when started early. But access to eye care isn’t always easy, especially in rural or low-income areas. So, developing effective, low-cost detection methods using deep learning is super promising. Models have shown high accuracy, sometimes even beating manual methods. But again, the lack of explainability is a major roadblock to getting these powerful tools into clinics for widespread screening.
Our Clever Solution: The DLMX Model
So, how did we tackle this? We came up with a methodology that really boosts medical explainability. We did it with two main innovative ideas:
- Multi-Task Learning Framework: We built a model that doesn’t just classify whether someone has AMD or not. It does that *and* it simultaneously performs lesion segmentation. Think of lesions like drusen, exudates, hemorrhages, and scars – the specific signs of AMD that doctors look for. By making the model identify and segment these lesions, it can essentially show you the evidence it used for its diagnosis. It’s like saying, “I diagnosed AMD because I see these specific drusen here.” This mirrors how clinicians make diagnoses!
- The Medical Explainability Index (MXI): This is our novel metric. We use Grad-CAM to generate a heatmap showing where the model looked for its classification decision. Then, the MXI measures how much that heatmap *overlaps* with the lesions the model segmented. It gives us a quantifiable way to say, “Okay, how much of the model’s decision is actually based on medically relevant features?” A high MXI means the model is focusing on the stuff doctors care about.
We call our proposed model Deep Learning with Medical eXplainability (DLMX). It’s not just about explaining; this multi-task approach actually helps the model perform better overall! By learning to classify AMD and segment lesions at the same time, the tasks help each other out. Segmentation gives detailed location info, while classification looks at the bigger picture. Sharing this info makes both tasks stronger.
How We Built It (The Nitty-Gritty, Briefly!)
Our DLMX model uses a U-Net architecture, which is great for image segmentation. It has an encoder (using powerful CNNs like EfficientNet or ResNet) to pull out features and a decoder to build the segmentation masks. The classification part takes features from the encoder. Both tasks are trained together using a combined loss function – a mix of binary cross-entropy for classification and cross-entropy plus Dice loss for segmentation. The Dice loss is super helpful because lesions can be small, and it handles that class imbalance nicely.
We initially focused on drusen because they’re the most common and defining feature of AMD. But the framework is designed to handle other lesions too, which we added later.
We trained and tested our model on the ADAM dataset, which has lots of fundus images with AMD labels and detailed annotations for different lesions. It’s a real-world dataset, complete with class imbalance (more non-AMD than AMD images), which we handled with resampling.
What the Numbers Tell Us
Alright, let’s talk results! We compared our DLMX model to single-task models (just classification or just segmentation) using the same backbone CNNs. For classification, we looked at metrics like accuracy, sensitivity, specificity, F1 score, and AUC (Area Under the Curve), which is great for imbalanced datasets. Our DLMX model, especially with EfficientNet-B7, performed really well, hitting an AUC of 0.96. It generally outperformed the single-task classification models.
For lesion segmentation, we used metrics like the Dice similarity coefficient (DSC) and Intersection over Union (IoU). DSC is awesome for small objects like lesions. Again, DLMX beat the single-task segmentation models, achieving a DSC of 0.59 for drusen segmentation. The multi-task approach clearly gives a boost, especially for the trickier segmentation task.
Why the improvement? As I mentioned, the tasks are related! Identifying drusen helps classify AMD, and knowing it’s an AMD image helps find the drusen. They learn from each other within the shared framework.
Putting the MXI to the Test
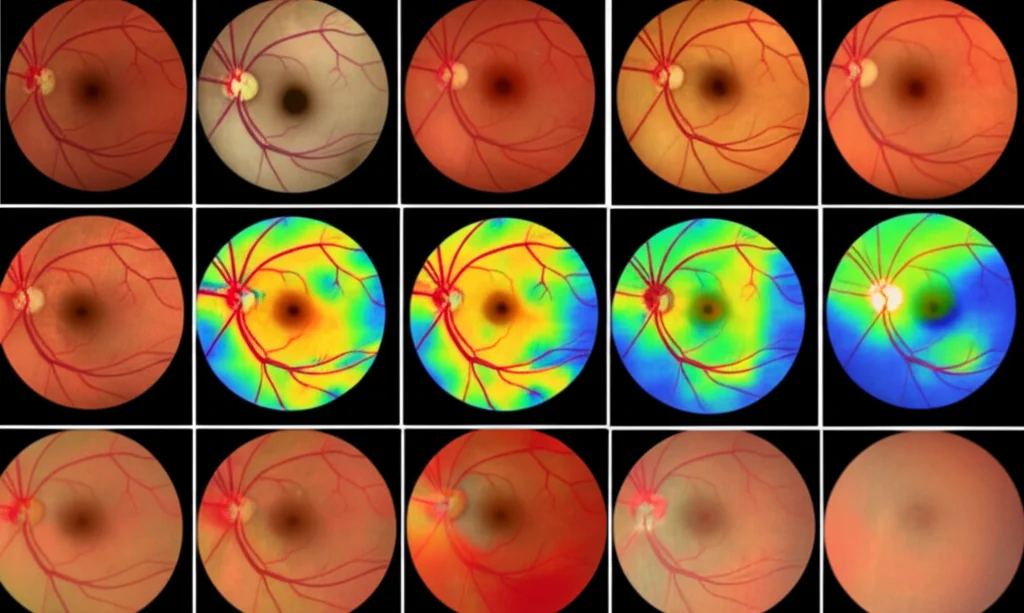
Now, for the star of the show: the Medical Explainability Index (MXI). Remember, this measures the overlap between the heatmap (where the AI looked) and the segmented lesion mask (what the AI identified as a lesion). We calculate it as an inclusion ratio – how much of the segmented lesion is covered by the heatmap.
We chose this inclusion ratio over something like DSC for MXI because we want to know if the model is looking at the lesion, even if it’s also looking at other stuff. DSC penalizes looking at extra areas, which isn’t what we want for explainability; we just want to know if the medically relevant area is *included* in the model’s focus.
To make sure our MXI was reliable, we also calculated a version using the expert-labeled ground truth lesion annotations (MXI_GT). The cool part? MXI (using the model’s own segmentation) and MXI_GT (using expert labels) showed a really strong positive correlation. This tells us that our model-generated MXI is a valid way to measure if the AI is focusing on medically relevant features, even without needing expert annotations every time – which is super practical for real-world use!
We saw examples where a high MXI (like 0.94) showed the heatmap strongly overlapped with the segmented drusen – meaning the model was basing its decision on the actual disease signs. In contrast, a low MXI (like 0.13) showed the heatmap focusing on other areas, like the optic disc, suggesting the decision wasn’t primarily based on the segmented drusen.
Beyond Drusen: Handling Multiple Lesions
We didn’t stop at just drusen! We expanded the model to segment other lesions too: exudates, hemorrhages, and scars. The model creates separate masks for each, then combines them into one big “composite lesion mask.” We then calculated MXI based on this combined mask. The results were consistent – MXI still showed a strong correlation with MXI_GT, even with the added complexity of multiple lesion types. This shows our framework is scalable and robust.
Balancing AI Power and Medical Knowledge
It’s a delicate balance, right? Deep learning is amazing at finding patterns we might not even know about. But medicine relies on established knowledge and known biomarkers. Our approach strikes this balance beautifully. By integrating lesion segmentation, we bake in that clinical knowledge (identifying known biomarkers) while the classification path lets the model learn other subtle patterns. The shared learning makes sure the model pays attention to the medically relevant stuff, leading to better performance and explainability.
Our results stack up well against others, even from competitions like the ADAM Challenge. Our DLMX model’s AUC for classification (0.96) and DSC for drusen segmentation (0.59) are competitive, and in the case of segmentation, better than the median competition results.
Interestingly, we noticed the heatmaps sometimes focused on the optic disc area, which has lots of blood vessels. This hints that the model might be picking up on vascular changes related to AMD, maybe even discovering new potential biomarkers! That’s exciting for future research.
Wrapping It Up
So, there you have it. We’ve developed a practical way to make deep learning models less of a black box for AMD diagnosis. By making the model identify the actual lesions it’s seeing and introducing the Medical Explainability Index (MXI) to quantify how much its decision relies on those lesions, we’re giving doctors and patients the transparency they need. This fosters trust and paves the way for AI to be more widely adopted in clinical practice, helping detect diseases like AMD earlier and more effectively. Plus, the framework is flexible enough to incorporate new biomarkers as we discover them. Pretty neat, huh?
Source: Springer